Dr. Sanjaya Abeysirigunawardena: "Modified nucleotides in ribosomal RNA and modification enzymes influence ribosomal assembly"

| Dr. Sanjaya Abeysirigunawardena Ph.D. Wayne State University, 2008 Associate Professor 126 Integrated Sciences Building sabeysir@kent.edu 330-672-2667 |
Study of Epitranscriptomic Regulation (funded by National Institutes of Health)
Understand the evolutionary importance of modified nucleotides in RNA-protein interactions during epitranscriptomic regulation.
- Investigate the key structural and sequence properties of proteins important for m6A recognition using phage display methods.
- Discover novel methyl-readers those involved in epitranscriptomic regulation using protein pulldown assays followed by bioinformatic analysis.
- Investigate key differences between direct and indirect readings of m6 A.
Drug Discovery
Discovery of small molecules that bind to various RNA drug targets.
- Discovery of novel antibacterial and anticancer agents by inhibiting RNA modification enzymes.
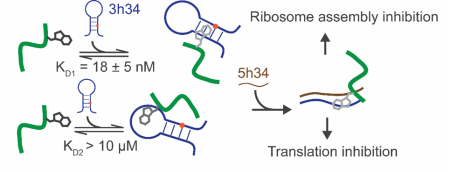
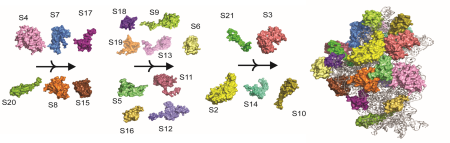
Ribosome Biogenesis
Investigate the ability of modification enzymes and their respective modifications to influence bacterial ribosomal assembly.
- Study the impact of modification enzymes on the co-transcriptional assembly of bacterial 30S ribosome subunit.
- Investigation of the ability of modification enzymes to modulate RNA dynamics using single molecule spectroscopy.
- Determination of the correlation of the antibiotic resistance in bacteria, ribosome assembly, and modification enzymes.
Meet the People in the Abey Research Group
Five graduate and over 30 undergraduate researchers have been actively involved in research in the Abey group. These undergraduate researchers include students majoring in Chemistry, Biology, and Biotechnology. In the Abey Lab, we strive for inclusivity!

To learn more about the Abey Research Group, please visit our website.
